HMMER Tutorial
HMMER: biosequence analysis using profile hidden Markov models
HMMER is used for searching sequence databases for homologs of protein sequences, and for making protein sequence alignments. It implements methods using probabilistic models called profile hidden Markov models (profile HMMs).
Compared to BLAST, FASTA, and other sequence alignment and database search tools based on older scoring methodology, HMMER aims to be significantly more accurate and more able to detect remote homologs because of the strength of its underlying mathematical models.
Take a look at the HMMER manual for more detailed information.
In this tutorial, we explain some of the features of the HMMER web application.
To begin, navigate to the HMMER web application via the menu bar, or go directly to http://hmmer.org/
Under the 'Organism menu', select one or several organisms that you would like to search against. Then, select which database should be searched. Unlike BLAST, available databases only include protein sequences predicted from the genome.
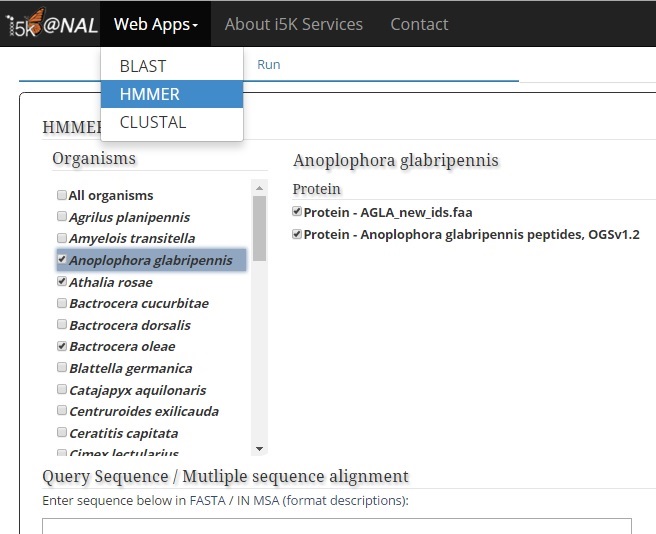
Paste your query sequence into the 'Query Sequence' field, or load it from disk.
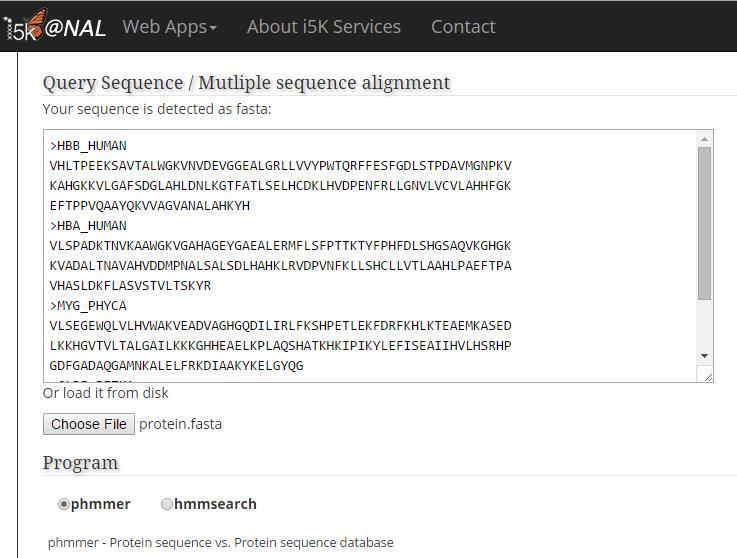
Program phmmer only accepts fasta files as query input, so it's not available when then input file is not in fasta format. However, the program hmmsearch accepts several different formats of multiple sequence alignment(which also include fasta). The format will be checked after submitting the query.
In this example, the query is in fasta format. Phmmer and Hmmsearch are both available.
In this example, the query is in CLUSTAL MSA format, and phmmer is greyed out (not available).
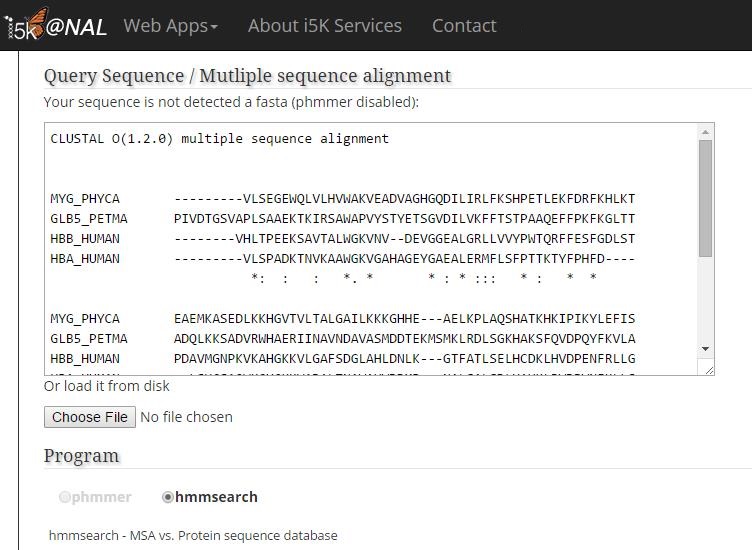
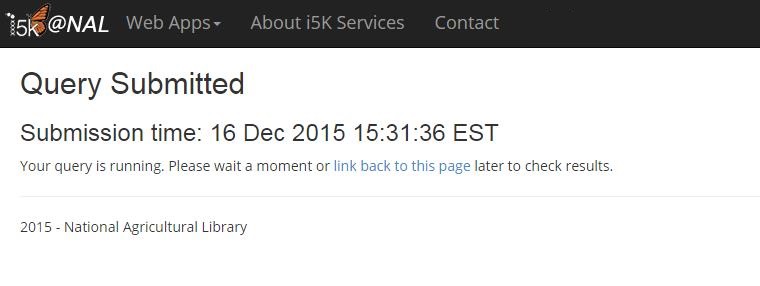
Once the correct HMMER program is selected, you can customize the parameters for your search. Hit the 'Search' button when finished.
A page will display indicating that your query is running. You can store the indicated query link if you wish to come back to your HMMER results at a later time. Results are stored for one week.
When the HMMER program has finished, your results will be displayed.
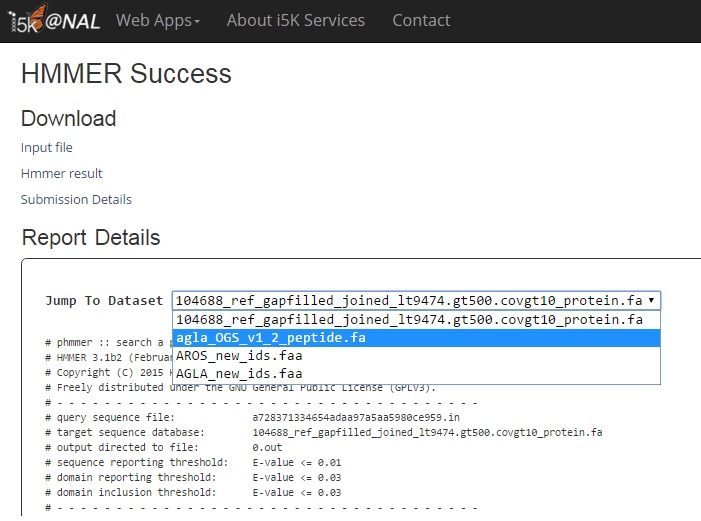
In 'Download' you can download the input file, the HMMER result (showed in 'Report Details') and the submission details including parameters used in this job.
In 'Report Details', reports are separated into independent sections by protein datasets. Use the drop-down list to navigate among them.