Rationale: Volunteer annotators manually improve a computationally predicted gene set from the i5k pilot project.
Before you begin:
- Identify the community contact of the organism you would like to help annotate from: Select your organism from the 'Organisms' drop-down menu on http://i5k.nal.usda.gov/. The community contact is listed under 'Organism details'.
- Communicate with your community contact about your annotation plans.
- Register to annotate your organism here: https://i5k.nal.usda.gov/web-apollo-registration.
- Study the Apollo development team's user guide to learn how to manually annotate in Apollo.
- Sign in to the Apollo installation for your organism using the registration information that was sent to you.
- Identify the computationally predicted gene set for your species that will serve as your 'starting point' for annotation. Ask your community contact or i5k@ars.usda.gov if you are unsure which track this is.
Rules: Once you have familiarized yourself with the manual annotation process in Apollo, follow these rules for manual annotation with the i5k pilot project:
-
Add information to a gene prediction.
- Add descriptive metadata about the gene prediction in the Information Editor (see below). Place all information in the 'mRNA' section of the Information Editor.
-
Modify an existing gene prediction.
- Perform gene structure modifications using the Web Apollo interface and external evidence. See the official Apollo user guide for more detailed examples. Only change an existing annotation if you have good external evidence.
- Add information about your modifications in the Information Editor (see below). Place all information in the 'mRNA' section of the Information Editor.
-
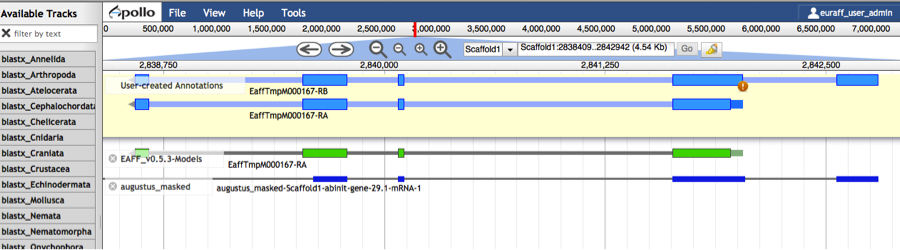
Create a new isoform.
- Perform gene structure modifications as for “2) Modify an existing gene prediction”. Name each new isoform with the suffix ‘-RB’, ‘-RC’, etc. (See example figure below).
- Additionally, elevate the original gene model/isoform to the user-created annotations track. If not, the original isoform will NOT be included in the final gene set.
-
Create a new gene prediction.
- We recommend only using existing evidence tracks (e.g. Augustus or Snap predictions) to generate your gene prediction.
- Add information to both the ‘gene’ and ‘mRNA’ sections of the Information Editor (see below).
-
Delete a gene prediction.
- Elevate the gene prediction to the User-created annotations track.
- In the Information Editor Status section, select the "Delete” radio button. Add information that supports your claim in the comments section.
The Information Editor.
- Name: This can be the full name of the gene plus isoform e.g. 'ultraspiracle isoform A'. Refer to our naming guidelines. For multiple isoforms, use the suffix “isoform A”, “isoform B”, etc
- Symbol: Add the gene symbol given sufficient evidence (e.g. 'USP').
- Description: A longer description of the mRNA name if necessary.
- Status: Click 'Approved' when you have confirmed that the annotation is correct. Click 'Delete' if this gene model should be deleted from the comptuationally predicted gene set.
- DBXRefs: Cross-references to other databases from which you have derived evidence, such as InterPro for protein domains, or UniProt for homology. Include the database name and evidence ID.
- PubMed IDs. If there is any published literature in support of this gene model, you can add the PubMed ID here.
- Gene Ontology IDs. Assign GO IDs if you have sufficient evidence for them.
- Comments. You may use the 'Comments' field to describe the actions you took for your annotation, and any other notes that you would like to communicate about the annotation.
 An official website of the United States government.
An official website of the United States government.